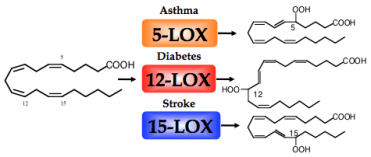
Introduction: Lipoxygenases are widely distributed throughout the plant and animal kingdoms and play a central role in their biology. In plants they are involved in germination and senescence. In mammalian tissue, there are three major human lipoxygenases (LOX), 5-LOX, 12-LOX, and 15-LOX, whose primary enzymatic difference is their positional specificity on arachidonic acid (AA). These lipoxygenase products are the precursors of important biomolecules, such as leukotrienes and lipoxins, which have been implicated as critical signaling molecules in a variety of inflammatory diseases. For example, 5-LOX is involved in asthma, 12-LOX in diabetes & atherosclerosis and 15-LOX in stroke & Alzheimer’s disease. These broad implications in human disease have elicited great interest in lipoxygenase as a potential therapeutic target and have motivated our lab to investigate lipoxygenase activity and inhibition.
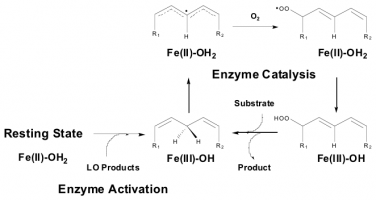
Lipoxygenase Enzyme Mechanism: In order to develop lipoxygenase inhibitors, one needs to understand its enzymatic function. Our lab achieves this by studying four lipoxygenase enzymes, soybean 15-LOX, human leukocyte 5-LOX, human platelet 12-LOX, human reticulocyte 15-LOX-1 and human epithelial 15-LOX-2 through a variety of spectroscopic and kinetic methods. The soybean enzyme is more robust than the human enzymes so it is used as a model system to develop a detailed mechanism for lipoxygenase as a class of enzyme. We have subsequently utilized this mechanism of soybean 15-LOX to compare with the human enzymes and determined that their mechanisms are remarkably similar despite their 100-fold difference in rate. We are now probing other LOX isozyme in order to understand the underlying principles of this varied class of enzymes.
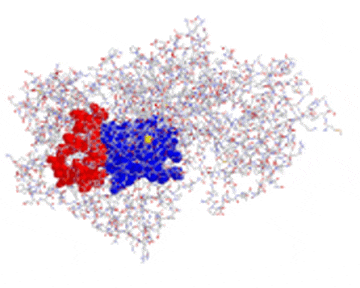
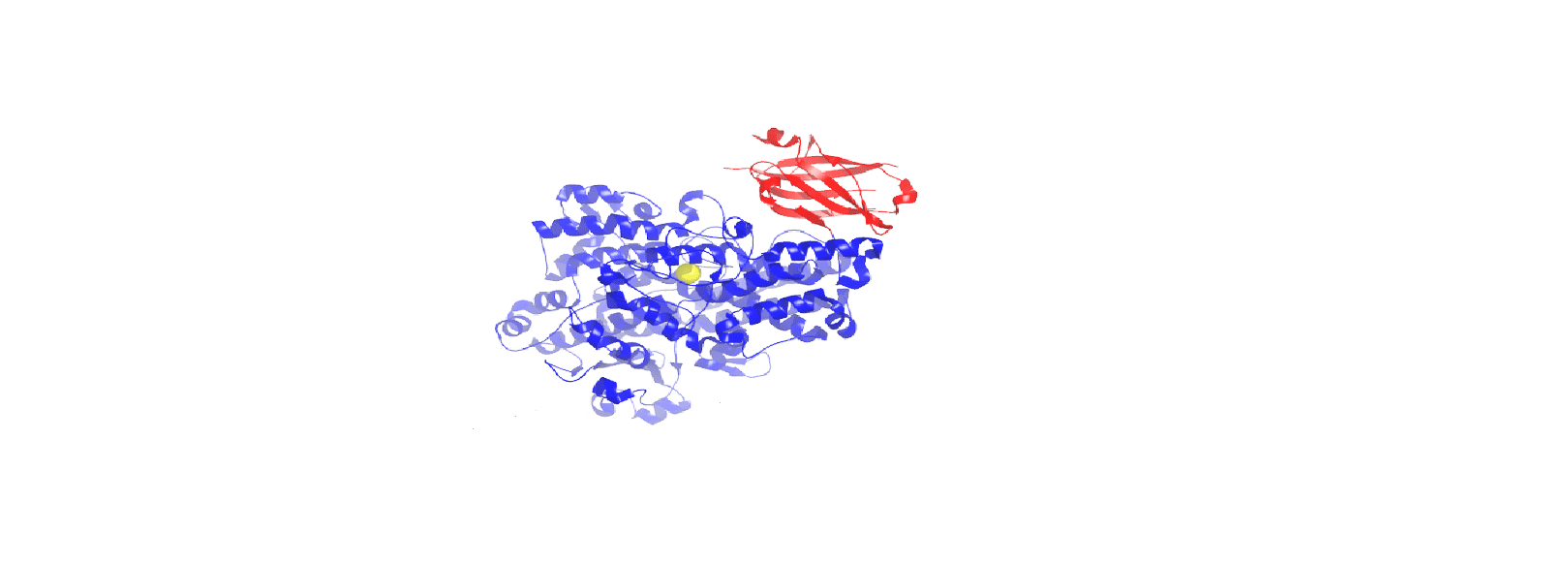
Lipoxygenase Allosteric Site: Our mechanistic investigations of LOX catalysis have helped us determine that many LOX isozymes have allosteric regulation sites. We believe that the allosteric site may be critical to the regulation of lipoxygenase activity by either suppressing or activating the enzyme. We also believe that the allosteric site could be used as an alternative site for inhibition and thus potentially be the target for a novel class of lipoxygenase inhibitors. We are currently probing the binding constraints of the allosteric site, which may help us to rationally design allosteric inhibitors.
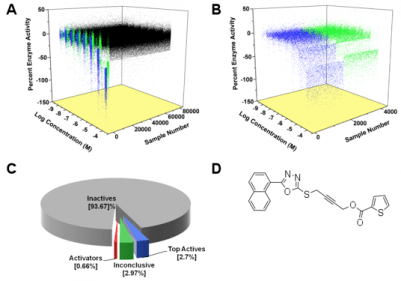
High Thru-Put Inhibitor Discovery: Our lab is also interested in the discovery and characterization of novel inhibitors to lipoxygenase. We currently have discovered over 50 unique lipoxygenase inhibitors through screening a marine natural products library (Prof. P. Crews) and the NIH Chemical Genomics Center (NCGC) inhibitor library. The library of Prof. Crews’ is one of the world’s largest marine natural products library and offers a unique source of novel chemical structures unavailable through standard synthetic or combinatorial methods. The NCGC library has over 400,000 compounds and was screened with advanced robotic methods in 8 hours, equivalent to 80 years by the manual method. We currently have hundreds of hits from both of these libraries and are investigating the most drug-like compounds with biochemical and spectroscopic methods to determine how they bind and inhibit lipoxygenase. For example, we are now investigating a 5-LOX inhibitor for anti-dandruff properties, a 12-LOX inhibitor for its anti-diabetic/ anti-atherosclerotic properties, and a 15-LOX-1 inhibitor for its anti-stroke/ anti-Alzheimer’s properties, with our biology collaborators.
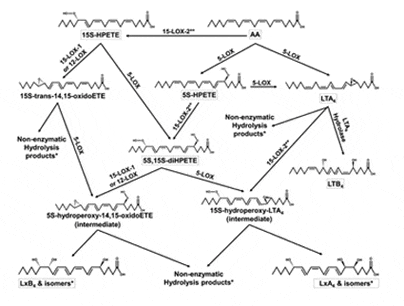
Oxy-Lipidomics via Mass Spectroscopy: In order to analyze the oxidized fatty acids in both our in vitro reactions and in vivo metabolic studies, we employ advanced mass spectroscopic methods. Our department has multiple mass spectrometers which allow us to identify trace oxy-lipids, which in turn instruct us in how LOX reacts with the greater than 30 fatty acids found in the human body. This information is critical because we have recently found that slight changes in oxy-lipid structure, such as length and/or unsaturation, can change the biological response as great as 10,000-fold. Therefore, identification of the specific bioactive oxy-lipid allows us to track it during in vivo LOX inhibition and possibly develop analogues for therapeutic use. In addition, we are determining with this lipidomic information that long held bio-synthetic pathways for these important oxy-lipid may not be correct. We are utilizing our specific LOX inhibitors to determine which LOX isozyme generates which oxy-lipid.